
Postdoctoral Researcher, molecular biologist Jacqueline Jerney working at the laboratory onboard research vessel Aranda. © Ilkka Lastumäki
The Finnish Environment Institute has tested the possibilities of using a genetic method to support the Finnish marine phytoplankton monitoring. The genetic method revealed two previously undetected species and one previously undetected genus in the northern Baltic Sea. According to the researchers, the combination of the genetic and microscopic methods would facilitate a better assessment of the total phytoplankton biodiversity and enable development of a biodiversity indicator in the future. The Finnish Environment Institute has published guidelines for the use of the eDNA method in marine phytoplankton monitoring.
Phytoplankton monitoring is conducted using the internationally standardized light microscopy method. In the GeMeKa project (link to the GeMeKa project´s website) funded by the Finnish Ministry of the Environment, researchers from the Finnish Environment Institute tested which additional information can be obtained by using the genetic method “environmental DNA (eDNA) metabarcoding” for phytoplankton monitoring. With eDNA metabarcoding, the entire DNA of a water sample can be analyzed, including free DNA and DNA inside of cells or colonies of many different organisms. The researchers analyzed the samples taken during the 2021 spring monitoring cruise onboard the marine research vessel Aranda using light microscopy and the eDNA method, combined the data, and compared the results obtained with the two methods. Based on the results, method guidelines were prepared for the use of eDNA technology in the Finnish marine phytoplankton monitoring.
The aim is to enhance biodiversity monitoring
The phytoplankton monitoring data are widely used in the assessment of the state of the Baltic Sea because the effects of eutrophication and climate change can be observed in the phytoplankton community often earlier than at the upper levels of the food web.
"Assessing phytoplankton biodiversity is challenging since taxonomic identification of microscopic cells is not simple. Currently, there is no HELCOM indicator for measuring phytoplankton biodiversity in the Baltic Sea area, although other types of phytoplankton indicators are already in use. There is a need for the development of a biodiversity indicator, as the biodiversity of phytoplankton significantly affects the production, stability and nutrient use efficiency in the pelagic ecosystems, which is reflected in the health of the entire marine ecosystem," says Senior Research Scientist Sirpa Lehtinen from the Finnish Environment Institute. Lehtinen coordinates the marine phytoplankton monitoring in Finland.
Genetic methods have the potential to increase information on phytoplankton biodiversity, especially of taxa which are impossible to identify microscopically (e.g. small-sized taxa), as well as of sparsely occurring taxa, which may go unnoticed with the standardized method.
However, the tested genetic method is not sufficient to replace the conventional microscopy-based monitoring method, because the genetic sequence databases lack a lot of Baltic Sea species, and the tested genetic method does not provide biomass data, which is needed for the calculation of eutrophication indicators and food web indicators used to assess the state of the Baltic Sea. However, combination of the genetic and microscopic methods would give a better assessment of the total phytoplankton biodiversity and enable development of a biodiversity indicator.
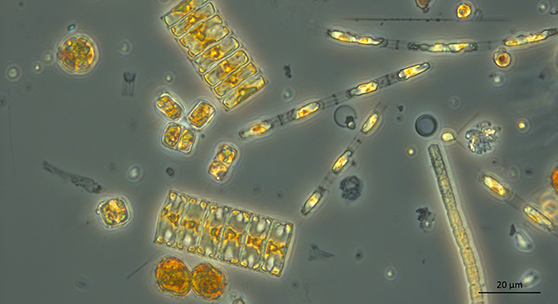
Phytoplankton under light microscope. The tested genetic method is not sufficient to replace the conventional microscopy-based monitoring method. However, combination of the genetic and microscopic methods would benefit the assessment of the total phytoplankton biodiversity. © Sirpa Lehtinen
New species were found in the northern Baltic Sea
Comparing the results of light microscopy and the eDNA method showed that some species were not detected with the eDNA method, but on the other hand, eDNA method also provided some information that was not obtained with light microscopy.
The eDNA method revealed two species new to the northern Baltic Sea: the cryptophyte Teleaulax gracilis and the diatom Thalassiosira hispida, as well as one new genus, the dinoflagellate genus Islandinium. In addition, the eDNA method detected the dinoflagellate Polarella glacialis, which is known to occur in the northern Baltic Sea, but only the resting cells of that species can be identified using the light microscope. In the western Gulf of Finland, the coccolithophorid family Noelaerhabdaceae was observed. Previously there were only uncertain observations of this family from the northern Baltic Sea. In addition, the eDNA method identified for example the single-celled picoplanktonic cyanobacterium genus Cyanobium, which cannot be identified with a light microscope.
Overall, there was a large diversity detected by the genetic method, but relatively few sequences were assigned to known species with the eDNA method. One reason is that many Baltic Sea species are not yet included in the genetic sequence databases, which are required as reference to assign sequences to species. "This result emphasizes the need to improve genetic sequence databases and optimize bioinformatics methods further," says Postdoctoral Researcher Jacqueline Jerney, the molecular biologist in the project.
Cooperation benefits the development of monitoring methods
Supplementing monitoring programs with genetic methods is a hot topic internationally and for example within the working groups of the Baltic Marine Environment Protection Commission HELCOM.
"At the EU level, we are aiming to restore our oceans and waters and one important part of this is to fill knowledge gaps and improve our understanding of marine and water ecosystems. One of the EU's goals is to sequence and openly publish 50 percent of the DNA of organisms inhabiting marine and aquatic ecosystems by 2030," says Postdoctoral Researcher Jacqueline Jerney.
National and international cooperation benefits the development of monitoring methods since that way knowledge and costs can be shared. During the GeMeKa project, the researchers of the Finnish Environment Institute organized five international online workshops, which were attended by experts in phytoplankton monitoring and genetic methods from the Baltic Sea region and the Nordic countries. The GeMeKa project cooperated with the Finnish Environment Institute's own seed money project, led by Researcher Tiina Laamanen, in the collection of samples and genetic analyzes of cyanobacteria.
The published method guidelines facilitate the implementation of eDNA technology
The method guidelines published by the Finnish Environment Institute contain detailed information on the collection of eDNA samples, sample processing, molecular biological work steps, bioinformatics analyses and quality assurance required for the high-throughput sequencing. The guidelines describe the use of 18S rRNA gene primers suitable for the identification of eukaryotic phytoplankton, and the 16S rRNA gene primers suitable for prokaryotic cyanobacteria. The guidelines will be further developed in the future as new research results, international guidelines and standards are published.
Inquiries:
- Senior Research Scientist Sirpa Lehtinen, e-mail: firstname.lastname@syke.fi
- Postdoctoral Researcher Jacqueline Jerney, e-mail: firstname.lastname@syke.fi